IMNN: Information maximising neural networks¶
The IMNN is a statistical method for transformation and compression of data from complex, high-dimensional distributions down to the number of physical parameters in the model which generates that data. It is asymptotically lossless in terms of information about the physical parameters. The method uses neural networks as a backbone for the transformation although any parameterised transformation with enough flexibility to map the data could be used.
Using simulations generated from the model the Fisher information (for a Gaussian distribution with parameter independent covariance) is calculated from the output of the transformation and its log determinant is maximised under the condition that the covariance of the outputted transformed simulations are approximately constant and approach the identity matrix.
Since the Fisher information only needs to be evaluated at a single fiducial parameter value it is exceptionally cheap to fit in comparison to other types of neural networks at the expense that the information is extracted from the data optimally about that fiducial parameter value and that the gradients of the simulations are needed (although this can be done numerically).
The ideal situation for performing inference is to fit an IMNN at a fiducial choice of parameters, evaluate the quasi-maximum-likelihood estimate of the parameters for the target data of choice and retrain a new IMNN at this estimate (still very cheap). This can be repeated iteratively until the IMNN is being trained at the maximum likelihood values of the parameters which will be most sensitive to the features in the data at that point and allow for the most precise constraints.
Note that the parameter estimates from the IMNN are not intended to be indicative of unbiased parameter estimates if the fiducial parameter choice is far from the maximum likelihood parameter values from some target. Instead they are meant to be used in a likelihood-free (simulation-based) inference scenario.
Using the IMNN¶
Implemented in JAX, the IMNN module provides a way to setup the fitting algorithm and fit an IMNN and then use it to make parameter estimates. There are several scenarios for the types of inputs that can be used.
SimulatorIMNN¶
Using a simulator (written in JAX or at least XLA compilable) for the generation of data provides the best results since generation of data is done on-the-fly. Since the focus of the IMNN is to maximise the information about the parameters from the data from the model, having a limited dataset means that spurious accidental correlations will almost certainly be learned about, overestimating the amount of information that can be extracted. For this reason, if using a limited sized training set like with GradientIMNN or NumericalGradientIMNN, then it is important to use a validation set and use early stopping based on the information extracted from this validation set. All these problems are side-stepped if simulations are generated on the fly since the same data is never seen multiple times. Of course, if the simulator is expensive then this may not be feasible.
The simulator needs to be of the form
def simulator(rng, parameters):
""" Simulate a realisation of the data
Parameters
----------
rng : int(2,)
A jax stateless random number generator
parameters : float(n_params,)
parameter values at which to generate the simulations at
Returns
-------
float(input_shape):
a simulation generated at parameter values `parameters`
"""
# Do stuff
return simulation
GradientIMNN¶
If simulations are expensive but a set of simulations with their gradients can be precalculated then it is possible to use these instead to train the IMNN. In this case the simulations are passed through the network and the Jacobian of the network outputs are calculated with respect to their inputs which using the chain rule can be combined with the gradient of the simulation with respect to the model parameters to get the Jacobian of the network outputs with respect to the physical model parameters, used for calculating the Fisher information. Although possible to not use a validation set when fitting it is highly recommended to use early stopping with a validation set to avoid overestimation of the amount of information extracted.
The simulations and their gradients should be of the form
import jax
from imnn.utils import value_and_jacrev
rng = jax.random.PRNGKey(0)
fiducial_parameters = # fiducial parameter values as a float(n_params,)
n_s = # number of simulations used to estimate covariance of network outputs
rng, *keys = jax.random.split(rng, num=n_s + 1)
fiducial, derivative = jax.vmap(
lambda key: value_and_jacrev(
simulator,
argnums=1)(key, fiducial_parameters))(
np.array(keys))
fiducial.shape
>>> (n_s, input_shape)
derivative.shape
>>> (n_s, input_shape, n_params)
Note that n_s derivatives are necessarily needed since only the mean of the derivatives is calculated which is more stable than the covariance. Therefore only n_d < n_s are required, although most stable optimisation is achieved using n_d = n_s.
NumericalGradientIMNN¶
If the gradient of the simulations with respect to the physical model parameters is not possible then numerical derivatives can be done. In this case simulations are made at the fiducial parameter value and then varied slightly with respect to each parameter independently with each of these simulations made at the same seed. Theses varied simulations are passed through the network and the outputs are used to make a numerical estimate via finite differences. There is quite a lot of fitting optimisation sensitivity to the choice of the finite difference size. Note that, again, it is VERY highly recommended to use a validation set for early stopping to prevent overestimation of the amount of information that can be extracted and the extraction of information from spurious features only existing in the limited dataset.
The simulations and their numerical derivatives should be made something like:
import jax
import jax.numpy as np
rng = jax.random.PRNGKey(0)
fiducial_parameters = # fiducial parameter values as a float(n_params,)
parameter_differences = # differences between varied parameter values for
# finite differences as a float(n_params,)
n_s = # number of simulations used to estimate covariance of network outputs
n_d = # number of simulations used to estimate the numerical derivative of
# the mean of the network outputs
rng, *keys = jax.random.split(rng, num=n_s + 1)
fiducial = jax.vmap(
lambda key: simulator(key, fiducial_parameters))(
np.array(keys))
varied_below = (fiducial_parameters - np.diag(parameter_differences) / 2)
varied_above = (fiducial_parameters + np.diag(parameter_differences) / 2)
below_fiducial = jax.vmap(
lambda key: jax.vmap(
lambda param: simulator(key, param))(varied_below))(
np.array(keys)[:n_d])
above_fiducial = jax.vmap(
lambda key: jax.vmap(
lambda param: simulator(key, param))(varied_above))(
np.array(keys)[:n_d])
derivative = np.stack([below_fiducial, above_fiducial], 1)
fiducial.shape
>>> (n_s, input_shape)
derivative.shape
>>> (n_s, 2, n_params, input_shape)
Matching seeds across pairs of varied parameters is fairly important for efficient training - stateless simulating like above makes this much easier.
Aggregation¶
If simulations or networks are very large then it can be difficult to fit an IMNN since the Fisher information requires the covariance to be well approximated to be able to maximise it. This means that all of the simulations must be passed through the network before doing a backpropagation step. To help with this, aggregation of computation and accumulated gradients are implemented. In this framework a list of XLA devices is passed to the IMNN class and data is passed to each device (via TensorFlow dataset iteration) to calculate the network outputs (and their derivatives using any of the SimulatorIMNN, GradientIMNN or NumericalGradientIMNN). These outputs are relatively small in size and so the gradient of the loss function (covariance regularised log determinant of the Fisher information) can be calculated easily. All of the data is then passed through the network again (a small number [n_per_device] at a time) and the Jacobian of the network outputs with respect to the neural network parameters is calculated. The chain rule is then used to combine these with the gradient of the loss function with respect to the network outputs to get the gradient of the loss function with respect to the network parameters. These gradients are summed together n_per_device at a time until a single gradient pytree for each parameter in the network is obtained which is then passed to the optimiser to implement the backpropagation. This requires two passes of the data through the network per iteration which is expensive, but is currently the only way to implement this for large data inputs which do not fit into memory. If the whole computation does fit in memory then there will be orders of magnitudes speed up compared to aggregation. However, aggregation can be done over as many XLA devices are available which should help things a bit. It is recommended to process as many simulations as possible at once by setting n_per_device to as large a value as can be handled. All central operations are computed on a host device which should be easily accessible (in terms of I/O) from all the other devices.
import jax
host = jax.devices("cpu")[0]
devices = jax.devices("gpu")
n_per_device = # number as high as makes sense for the size of data
TensorFlow Datasets¶
If memory is really tight and data needs to be loaded from disk then it is possible to use TensorFlow Datasets to do this, but the datasets must be EXTREMELY specifically made. There are examples in the examples directly, but shortly there are two different variants, the DatasetGradientIMNN and the DatasetNumericalGradientIMNN. For the DatasetNumericalGradientIMNN the datasets must be of the form
import tensorflow as tf
fiducial = [
tf.data.TFRecordDataset(
sorted(glob.glob("fiducial_*.tfrecords")),
num_parallel_reads=1
).map(writer.parser
).skip(i * n_s // n_devices
).take(n_s // n_devices
).batch(n_per_device
).repeat(
).as_numpy_iterator()
for i in range(n_devices)]
derivative = [
tf.data.TFRecordDataset(
sorted(glob.glob("derivative_*.tfrecords")),
num_parallel_reads=1
).map(writer.parser
).skip(i * 2 * n_params * n_d // n_devices
).take(2 * n_params * n_d // n_devices
).batch(n_per_device
).repeat(
).as_numpy_iterator()
for i in range(n_devices)]
Here the tfrecords contains the simulations which are parsed by the writer.parser (there is a demonstration in imnn.TFRecords). The simulations are split into n_devices different datasets each which contain n_s // n_devices simulations which are passed to the network n_per_device at a time and repeated and not shuffled. For derivative, because there are multiple simulations at each seed for the finite differences then 2 * n_params * n_d // n_devices need to be available to each device before passing n_per_device to the network on each device.
For the DatasetGradientIMNN the loops are made quicker by separating the derivative and simulation calculation from the simulation only calculations (the difference between n_s and n_d). In this case the datasets must be constructed like:
fiducial = [
tf.data.TFRecordDataset(
sorted(glob.glob("fiducial_*.tfrecords")),
num_parallel_reads=1
).map(writer.parser
).skip(i * n_s // n_devices
).take(n_s // n_devices)
for i in range(n_devices)]
main = [
tf.data.Dataset.zip((
fiducial[i],
tf.data.TFRecordDataset(
sorted(glob.glob("derivative_*.tfrecords")),
num_parallel_reads=1).map(
lambda example: writer.derivative_parser(
example, n_params=n_params)))
).take(n_d // n_devices
).batch(n_per_device
).repeat(
).as_numpy_iterator()
for i in range(n_devices)]
remaining = [
fiducial[i].skip(n_d // n_devices
).take((n_s - n_d) // n_devices
).batch(n_per_device
).repeat(
).as_numpy_iterator()
for i in range(n_devices)]
Note that using datasets can be pretty tricky, aggregated versions of GradientIMNN and NumericalGradientIMNN does all the hard work as long as the data can be fit in memory.
Neural models¶
The IMNN is designed with stax-like models and jax optimisers which are very flexible and designed to be quickly developed. Note that these modules don’t need to be used exactly, but they should look like them. Models should contain
def initialiser(rng, input_shape):
""" Initialise the parameters of the model
Parameters
----------
rng : int(2,)
A jax stateless random number generator
input_shape : tuple
The shape of the input to the network
Returns
-------
tuple:
The shape of the output of the network
pytree (list or tuple):
The values of the initialised parameters of the network
"""
# Do stuff
return output_shape, initialised_parameters
def apply_model(parameters, inputs):
""" Passes inputs through the network
Parameters
----------
parameters : pytree (list or tuple)
The values of the parameters of the network
inputs : float(input_shape)
The data to put through the network
Returns
-------
float(output_shape):
The output of the network
"""
# Do neural networky stuff
return output
model = (initialiser, apply_model)
The optimiser also doesn’t specifically need to be a jax.experimental.optimizer, but it must contain
def initialiser(initial_parameters):
""" Initialise the state of the optimiser
Parameters
----------
parameters : pytree (list or tuple)
The initial values of the parameters of the network
Returns
-------
pytree (list or tuple) or object:
The initial state of the optimiser containing everything needed to
update the state, i.e. current state, the running mean of the
weights for momentum-like optimisers, any decay rates, etc.
"""
# Do stuff
return state
def updater(it, gradient, state):
""" Updates state based on current iteration and calculated gradient
Parameters
----------
it : int
A counter for the number of iterations
gradient : pytree (list or tuple)
The gradients of the parameters to update
state : pytree (list or tuple) or object
The state of the optimiser containing everything needed to update
the state, i.e. current state, the running mean of the weights for
momentum-like optimisers, any decay rates, etc.
Returns
-------
pytree (list or tuple) or object:
The updated state of the optimiser containing everything needed to
update the state, i.e. current state, the running mean of the
weights for momentum-like optimisers, any decay rates, etc.
"""
# Do updating stuff
return updated_state
def get_parameters(state):
""" Returns the values of the parameters at the current state
Parameters
----------
state : pytree (list or tuple) or object
The current state of the optimiser containing everything needed to
update the state, i.e. current state, the running mean of the
weights for momentum-like optimisers, any decay rates, etc.
Returns
-------
pytree (list or tuple):
The current values of the parameters of the network
"""
# Get parameters
return current_parameters
optimiser = (initialiser, updater, get_parameters)
IMNN¶
Because there are many different cases where we might want to use different types of IMNN subclasses. i.e. with a simulator, aggregated over GPUs, using numerical derivatives, etc. then there is a handy single function will try and return the intended subclass. This is
import imnn
IMNN = imnn.IMNN(
n_s, # number of simulations for covariance
n_d, # number of simulations for derivative mean
n_params, # number of parameters in physical model
n_summaries, # number of outputs from the network
input_shape, # the shape a single input simulation
θ_fid, # the fiducial parameter values for the sims
model, # the stax-like model
optimiser, # the jax optimizers-like optimiser
key_or_state, # either a random number generator or a state
simulator=None, # SimulatorIMNN simulations on-the-fly
fiducial=None, # GradientIMNN or NumericalGradientIMNN sims
derivative=None, # GradientIMNN or NumericalGradientIMNN ders
main=None, # DatasetGradientIMNN sims and derivatives
remaining=None, # DatasetGradientIMNN simulations
δθ=None, # NumericalGradientIMNN finite differences
validation_fiducial=None, # GradientIMNN or NumericalGradientIMNN sims
validation_derivative=None, # GradientIMNN or NumericalGradientIMNN ders
validation_main=None, # DatasetGradientIMNN sims and derivatives
validation_remaining=None, # DatasetGradientIMNN simulations
host=None, # Aggregated.. host computational device
devices=None, # Aggregated.. devices for running network
n_per_device=None, # Aggregated.. amount of data to pass at once
cache=None, # Aggregated.. whether to cache simulations
prefetch=None,) # Aggregated.. whether to prefetch sims
So for example to initialise an AggregatedSimulatorIMNN and train it we can do
rng, key = jax.random.split(rng)
IMNN = imnn.IMNN(n_s, n_d, n_params, n_summaries, input_shape,
fiducial_parameters, model, optimiser, key,
simulator=simulator, host=host, devices=devices,
n_per_device=n_per_device)
rng, key = jax.random.split(rng)
IMNN.fit(λ=10., ϵ=0.1, rng=key)
IMNN.plot(expected_detF=50.)
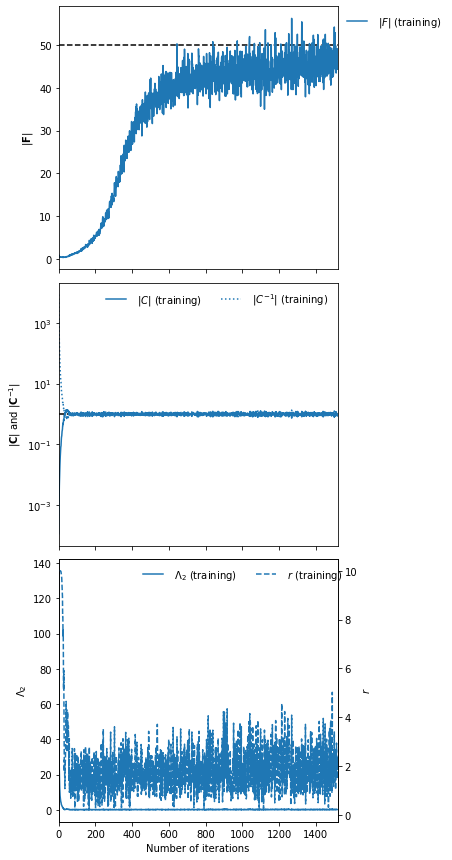
Or for a NumericalGradientIMNN
rng, key = jax.random.split(rng)
IMNN = imnn.IMNN(n_s, n_d, n_params, n_summaries, input_shape,
fiducial_parameters, model, optimiser, key,
fiducial=fiducial, derivative=derivative,
δθ=parameter_differences,
validation_fiducial=validation_fiducial,
validation_derivative=validation_derivative)
IMNN.fit(λ=10., ϵ=0.1)
IMNN.plot(expected_detF=50.)
λ and ϵ control the strength of regularisation and should help with speed of convergence but not really impact the final results.
Citing work¶
If you use this code please cite
@article{charnock2018,
author={Charnock, Tom and Lavaux, Guilhem and Wandelt, Benjamin D.},
title={Automatic physical inference with information maximizing neural networks},
volume={97},
ISSN={2470-0029},
url={http://dx.doi.org/10.1103/PhysRevD.97.083004},
DOI={10.1103/physrevd.97.083004},
number={8},
journal={Physical Review D},
publisher={American Physical Society (APS)},
year={2018},
month={Apr}
}
and maybe also
@software{imnn2021,
author = {Tom Charnock},
title = {{IMNN}: Information maximising neural networks},
url = {http://bitbucket.org/tomcharnock/imnn},
version = {0.3.0},
year = {2021},
}