Using the AggregatedSimulatorIMNN¶
ith a generative model built in jax we can generate simulations
on-the-fly to fit an IMNN. Furthermore, if the simulations are too
numerous or too large to fit into memory or could be accelerated over
several devices, we can aggregate the gradients too. This is an
expensive operation and should only be used if memory over a single
device is really an issue. The SimulatorIMNN (and its aggregated form)
are the best way to prevent against overfitting due to limited
variability in datasets which may introduce spurious features which are
not informative about the model parameters.
For this example we are going to summaries the unknown mean, \(\mu\), and variance, \(\Sigma\), of \(n_{\bf d}=10\) data points of two 1D random Gaussian field, \({\bf d}=\{d_i\sim\mathcal{N}(\mu,\Sigma)|i\in[1, n_{\bf d}]\}\). This is an interesting problem since we know the likelihood analytically, but it is non-Gaussian
As well as knowing the likelihood for this problem, we also know what sufficient statistics describe the mean and variance of the data - they are the mean and the variance
What makes this an interesting problem for the IMNN is the fact that the sufficient statistic for the variance is non-linear, i.e. it is a sum of the square of the data, and so linear methods like MOPED would be lossy in terms of information.
We can calculate the Fisher information by taking the negative second derivative of the likelihood taking the expectation by inserting the relations for the sufficient statistics, i.e. and examining at the fiducial parameter values
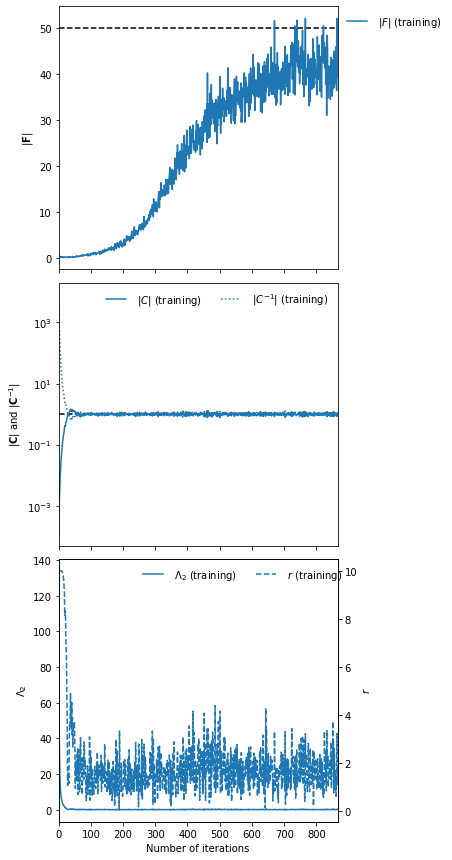
Choosing fiducial parameter values of \(\mu^\textrm{fid}=0\) and \(\Sigma^\textrm{fid}=1\) we find that the determinant of the Fisher information matrix is \(|{\bf F}_{\alpha\beta}|=50\).
from imnn import AggregatedSimulatorIMNN
import jax
import jax.numpy as np
from jax.experimental import stax, optimizers
We’re going to use 1000 summary vectors, with a length of two, at a time to make an estimate of the covariance of network outputs and the derivative of the mean of the network outputs with respect to the two model parameters.
n_s = 1000
n_d = 100
n_params = 2
n_summaries = n_params
input_shape = (10,)
The simulator is simply
def simulator(key, θ):
return θ[0] + jax.random.normal(key, shape=input_shape) * np.sqrt(θ[1])
Our fiducial parameter values are \(\mu^\textrm{fid}=0\) and \(\Sigma^\textrm{fid}=1\).
θ_fid = np.array([0., 1.])
For initialising the neural network and for generating the simulations on the fly we need two random number generators:
rng = jax.random.PRNGKey(1)
rng, model_key, fit_key = jax.random.split(rng, num=3)
We’re going to use jax’s stax module to build a simple network
with three hidden layers each with 128 neurons and which are activated
by leaky relu before outputting the two summaries. The optimiser will be
a jax Adam optimiser with a step size of 0.001.
model = stax.serial(
stax.Dense(128),
stax.LeakyRelu,
stax.Dense(128),
stax.LeakyRelu,
stax.Dense(128),
stax.LeakyRelu,
stax.Dense(n_summaries))
optimiser = optimizers.adam(step_size=1e-3)
We will use the CPU as the host memory and use the GPUs for calculating the summaries.
host = jax.devices("cpu")[0]
devices = jax.devices("gpu")
Now lets say that we know that we can process 100 simulations at a time per device before running out of memory, we therefore can set
n_per_device = 100
The AggregatedSimulatorIMNN can now be initialised setting up the network and the fitting routine (as well as the plotting function)
imnn = AggregatedSimulatorIMNN(
n_s=n_s, n_d=n_d, n_params=n_params, n_summaries=n_summaries,
input_shape=input_shape, θ_fid=θ_fid, model=model,
optimiser=optimiser, key_or_state=model_key, host=host,
devices=devices, n_per_device=n_per_device,
simulator=simulator)
To set the scale of the regularisation we use a coupling strength \(\lambda\) whose value should mean that the determinant of the difference between the covariance of network outputs and the identity matrix is larger than the expected initial value of the determinant of the Fisher information matrix from the network. How close to the identity matrix the covariance should be is set by \(\epsilon\). These parameters should not be very important, but they will help with convergence time.
λ = 10.
ϵ = 0.1
Fitting can then be done simply by calling:
imnn.fit(λ, ϵ, rng=fit_key, print_rate=1)
0it [00:00, ?it/s]
Here we have included a print_rate for a progress bar, but leaving
this out will massively reduce fitting time (at the expense of not
knowing how many iterations have been run). The IMNN will be fit for a
maximum of max_iterations = 1000 iterations, but with early stopping
which can turn on after min_iterations = 100 iterations and after
patience = 10 iterations where the maximum determinant of the Fisher
information matrix has not increased. imnn.w is set to the values of
the network parameters which obtained the highest value of the
determinant of the Fisher information matrix, but the values at the
final iteration can be set using best = False.
To continue training one can simply rerun fit
imnn.fit(λ, ϵ, patience=10, max_iterations=1000, print_rate=1)
although we will not run it in this example.
To visualise the fitting history we can plot the results:
imnn.plot(expected_detF=50);